Our inaugural blog post is recapitulating our bi-weekly journal club. We came together and discussed the paper “Hierarchical clustering of gene-level association statistics reveals shared and differential genetic architecture among traits in the UK Biobank” by Melissa R. McGuirl and colleagues.
What is the paper about:
Introduction of a new hierarchical clustering algorithm called WINGS for gene-based phenotype associations between many traits to identify phenotypes with shared core genes.
The paper:
The new method:
The authors used a previously released software tool by the same group called PEGASUS to calculate gene level p-values for disease associations for many different traits.
These gene level p-values were then used as feature vectors for the here newly proposed clustering pipeline called WINGS (implemented in MATLAB).
First Ward hierarchical clustering was applied to the feature vector. This analysis will identify traits which are highly similar based on gene phenotype associations and group them together in a cluster. In the next step these cluster definitions are ranked and scored regarding their importance.
Dendograms can be used to visualize the clustering:
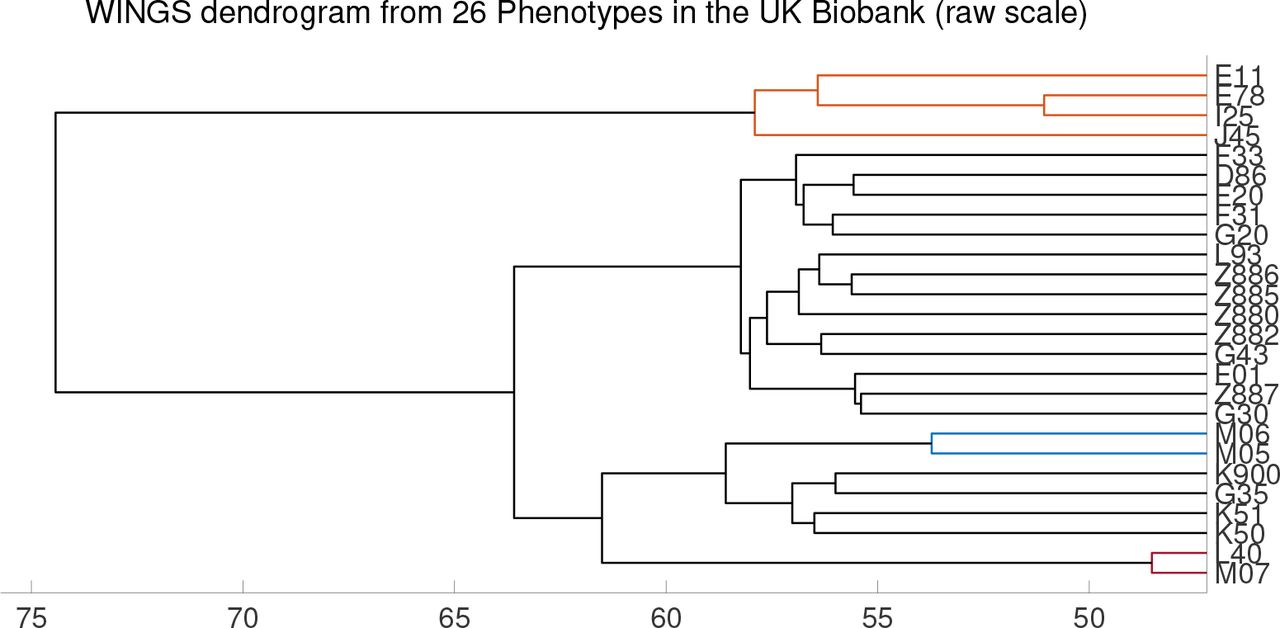
Based on the length of branches in these dendograms, the authors then identify “significant clusters” by filtering for maximum outliers in the distribution of branch lengths. These “significant clusters” are thought to contain traits which share core genetic risk factors.
WINGS as well as PEGASUS are being made available on github.
Data sets used:
The authors first used simulated data sets to measure the performance of WINGS and later applied the pipeline to 87 case-control phenotypes in the UK biobank.
Caveats:
A few caveats of this study became apparent during our discussion of the methods and results in the journal club. First, the generation of single gene trait association p values is not trivial. The authors state variants in a window of +/−50kb around autosomal genes were considered for the association analysis. This can lead to clumping of signals for overlapping genes or genes in close proximity.
Second, the designation of clusters as “significant clusters” based on the arbitrary branch length threshold chosen is a little misleading.
And third, the sample sizes and statistical power of the underlying genome-wide association studies used to generate the gene level p values will effect the analysis and results. For future versions of the pipeline a normalization of p values based on the effective sample size could be considered.
Innovation:
The here published method presents a compelling combination of burden testing and clustering based on gene trait associations. The applied clustering allows to identify groups of traits with higher priority and presumably closer overlap in genetic architecture. This method offers a range of attractive future applications that could provide further insights into the genetic architecture. A possible application is the analysis of the genetic architecture for traits between different ancestry groups allowing an examination independent of varying variant frequencies and LD structures.
