Influences of copy number variants (CNVs) and polygenic risk scores (PRSs) in psychotic disorders: A prospective study and meta-analysis
Over 100 single nucleotide polymorphisms (SNPs) are associated with psychosis. Individually their predictive power is relatively small, and due to the polygenecity of psychosis their combination to calculate a polygenic risk score (PRS) has been proposed as a better alternative. Several large and rare copy number variants (CNVs), which are either deletions or duplications of DNA sequence, resulting in a copy number other than two, has also been reported to increase the risk of developing psychosis.
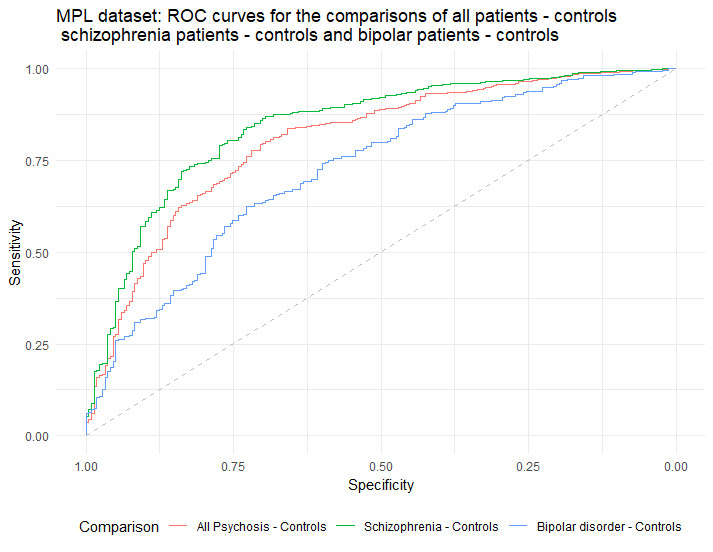
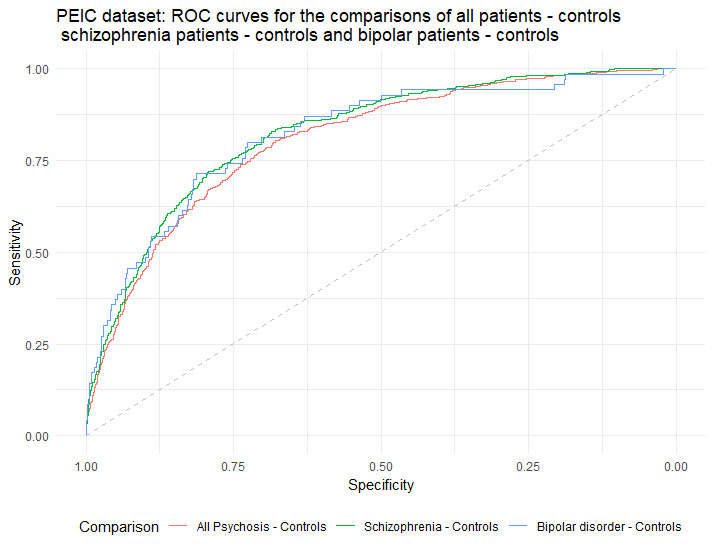
In this project we are analysing two large UCL samples: the Molecular Psychiatry Lab from UCL (MPL=1,266) and the Psychosis Endophenotypes Consortium (PEIC=2,429) samples, resulting in a total of 1,302 schizophrenia patients, 348 patients with bipolar disorder, 100 patients with other psychotic disorders and 1,945 healthy controls.
We aim to investigate whether CNV burden, as measured by number of genes affected, and Polygenic Risk Scores for both schizophrenia and bipolar disorder with psychotic features, could distinguish between patients with psychosis (either schizophrenia or bipolar with psychotic features) and healthy controls. We then produce Receiver Operation Characteristic (ROC) curves, and meta-analyse the Areas Under the Curve (AUC).
A better understanding of PRS and CNVs influences on the risk of developing psychosis is crucial for developing new treatments and could be useful towards early detection and treatment of psychosis, which leads to better outcomes and prognosis.
Collaborators
- Dr. Johan Thygesen
- Irina Zartaloudi
- Dr. Haritz Irizar
- Dr. Karoline Kuchenbaecker
- Dr. Olga Giannakopoulou
- Dr. Stefanie Mueller
- Dr. Stella Calafato
- Isabelle Austin-Zimmerman
- Anjali Bhat
- Jasmine Harju-Seppänen, Division of Psychiatry, University College London, London, United Kingdom
- Rebecca Muir, Division of Psychiatry, University College London, London, United Kingdom
- Prof. Andrew McQuillin
- Prof. Elvira Bramon