This blog post summarizes one of our bi-weekly journal club discussion of the paper titled “Delineating the Genetic Component of Gene Expression in Major Depression” by Lorenza Dall’Aglio, Cathryn M. Lewis and Oliver Pain. The group works in the department of Medical and Molecular Genetics, Faculty of Life Sciences and Medicine at King’s College London.
The paper:
Aim of the paper:
To shed light on the genetic component of genetic expression in major depression (MD) through the functional characterization of previously associated genes, the identification of new risk loci and the identification of functionally enriched pathways.
Methods & Datasets used:
The authors performed a Transcription-Wide Association Study (TWAS) to identify differentially expressed genes in MD. They used the TWAS FUSION method (Gusev et al., 2016) that uses only summary statistics. The authors used summary statistics from one of the latest and largest GWAS for MD by Wray et al., (2018), which considered 135,458 MD cases and 344,901 controls. They also used SNP-weights from a range of tissues, including whole blood, peripheral blood, adrenal, pituitary and thyroid glands. The SNP-weights represent the estimated correlations of the effect of SNPs with the expression of their annotated gene. For this study, 20 SNP-weight sets were extracted from four RNA reference datasets: the Netherlands Twins Register (NTR), the Young Finns Study (YFS), the CommonMind Consortium (CMC) and finally, the Genotype-Tissue Expression (GTEx) Consortia. Finally, authors used the 1000 Genomes Phase 3 European reference panel for LD estimation.
Findings:
This study identified 177 transcriptome wide significant associations for 91 unique genes across all SNP-weight sets as depicted in Figure 1.
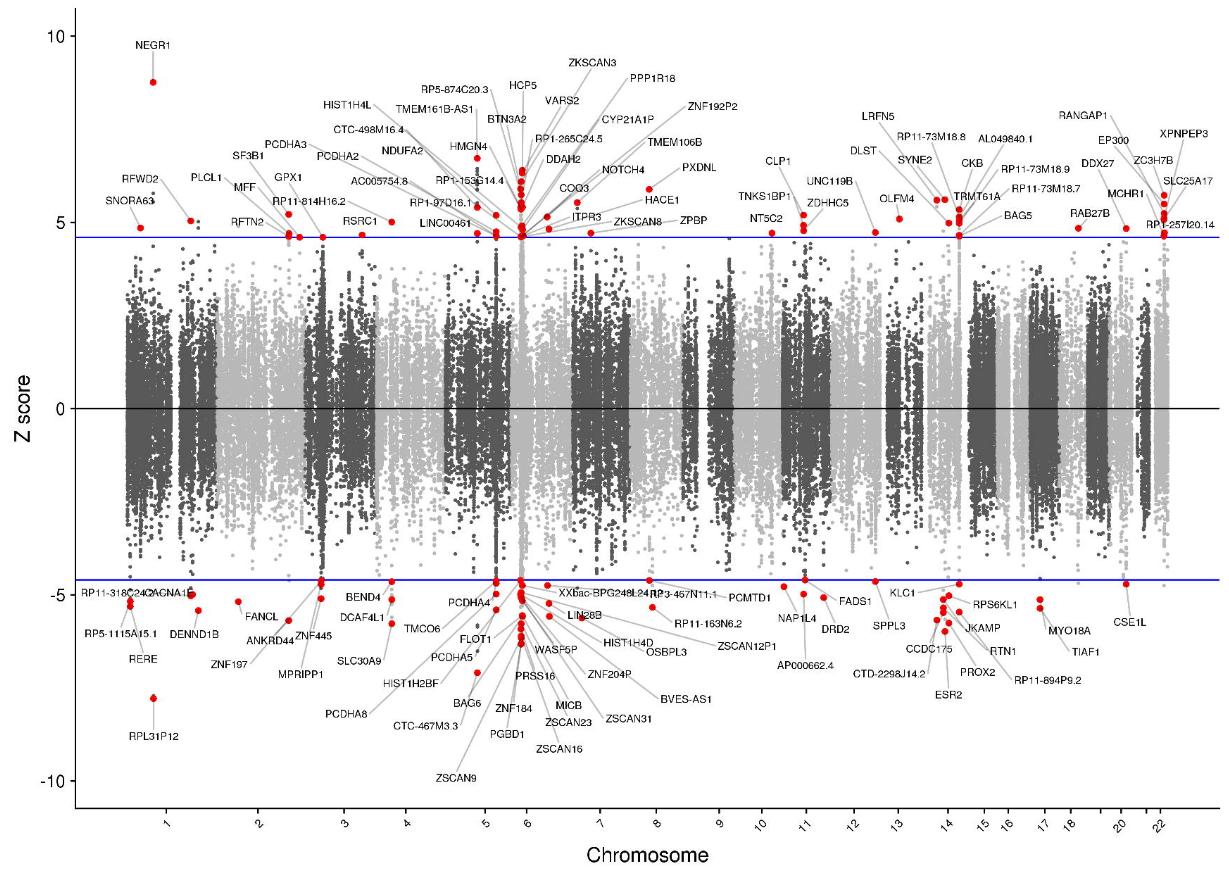
Of these genes, 50 were novel, undetected by previous TWASs, 41 were outside GWAS-significant loci. Fifty-seven of the significant genes were colocalized, 44 were independent, and 8 were identified as high-confidence associations based on strong TWAS association, colocalization and causal fine-mapping. From the 8 high-confidence associated genes found, 3 have intriguing functional characteristics. NEGR1 is implicated in axon extension and synaptic plasticity, ESR2 regulates estrogen activity and TMEM106B was highlighted as a susceptible gene for neurodegenerative disorders.
Innovations:
The authors included an abundance of tissues and added extra steps in the traditional TWAS analysis, including colocalization, conditional and fine-mapping analysis, which led to new findings associated with major depression. The fact that they used previously published data to unravel new gene associations with the disease shows once again that data mining through different techniques can lead to additional discoveries.
Caveats:
Some limitations of the study, such as the small sample sizes of the gene expression reference samples and the possible noise introduced by the wider set of tissues have been discussed by the authors. However, it was not clear in the text how the authors took into account the different tissue when they adjusted for multiple testing. We also extensively discussed the additional information that some follow-up analyses can contribute to main TWAS results. For example, the authors conducted colocalization analysis to estimate the probability that associations within a locus for both gene expression and MD are driven by a shared causal variant. This could benefit from more details as we thought that this is, at least in some extent, considered in the FUSION analysis. Furthermore, it was not clear which biological processes or pathways the sets are capturing, with some exceptions such as the set labelled “ALL.ION.CHANNELS”. A more detailed description of the pathways would be helpful for the reader.
